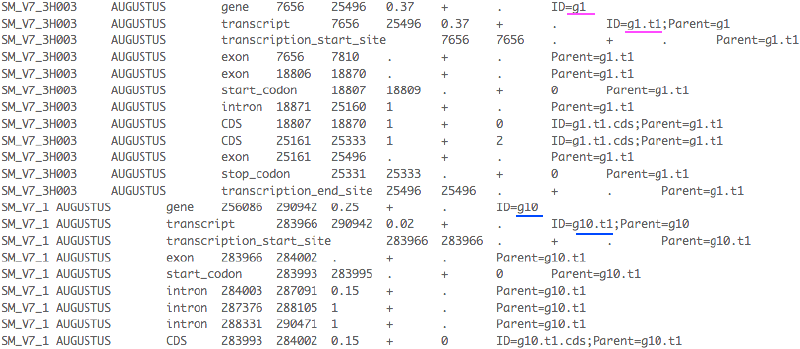
I’ve recently been doing identifier transfer work for the parasitic worm Schistosoma mansoni. We have two gene sets: the RATT transferred set from the old annotations, and the Augustus set with predicted annotations. The agreement is to keep the features in Augustus, but take the identifiers from RATT.
One approach I thought about is firstly to generate a confident identifier pair list (key-value pairs) like this:
|
|
Then use text replacement to change the IDs in Augustus with their RATT counterparts, but pay attention not to replace the string (like g1) in another string (like g10); and we wanted to keep the naming of transcript like “xxx.1” instead of “xxx.t1”.
I tried to use Bash associative array like what I did for pairwise sequence alignment, but I did not get any success. Then I relized that Perl has Hash and Python has Dictionary, and there might be script already available. Finally I came to a script like this (sadly at this time point I cannot find the original post) with a bit modifications:
|
|
Bingo!

I made a first try and it took 11 hours to replace 13,300 string pairs.
UPDATE 26.05.2017 Changes were made in the script to make exact replacement (not to mis-replace “g1” in “g10”).