As we know microarray deals with probes.
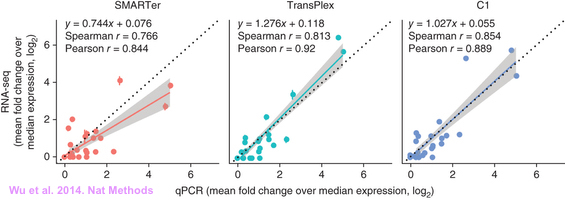
One problem is that in a single experiment the limited number of hybridization probes can hardly represent the whole transcript population in the sample. This would result in a probe bias and reduced linear range. So the standard procedure is to technically validate microarray results by qPCR. However, as RNA-seq detects the entire transcripts and through a series of quality control and normalization the technical bias is greatly reduced. On the other hand, qPCR has its own prob bias. Personally I have not seen many people include qPCR in their RNA-seq paper. But for those who did qPCR validation, they found good correlation between the two approaches.
Weather qCPR is really necessary for RNA-Seq, here are people’s opinions:
We had considered performing qPCR studies to ‘re-validate’ some of our gene-expression findings but there is little evidence that qPCR analyses from the same samples will add any extra utility to our data so we decided to eschew those experiments. – Dave Bridges
In my opinion, the validation by qPCR was not essential, but is useful. – a paper reviewer.
Validating your experiment is not that necessary I believe; precising an observation is nice on the other hand. Usually, I see articles comparing micro-array and RNASeq data, not RNA Seq and RT-qPCR. – a Biostar thread
Although validation is required by a lot of journals for publication, it is still debatable whether qRT-PCR validation of differentially expressed genes is still necessary…and how to do it exactly if the answer is yes. – Design and validation issues in RNA-seq experiments
In summary qPCR could be more helpful if no biological replicates were used in RNA-Seq. However, if the reviewer or the journal requires such validation even if replicates are indicated, it’s out of this discussion and you just need to do it.