Glossary
- CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)
- Cas (CRISPR-associated genes)
- crRNA (CRISPR RNA)
- traceRNA/trRNA (trans-activating crRNA)
- gRNA (guide RNA)
- sgRNA (single guide RNA)
- Indel (insertion and/or deletion)
- PAM (Protospacer-Adjacent Motif)
- ZNF (Zinc-Finger Nuclease)
 {:hight=“348px” width=“550px”}
{:hight=“348px” width=“550px”}
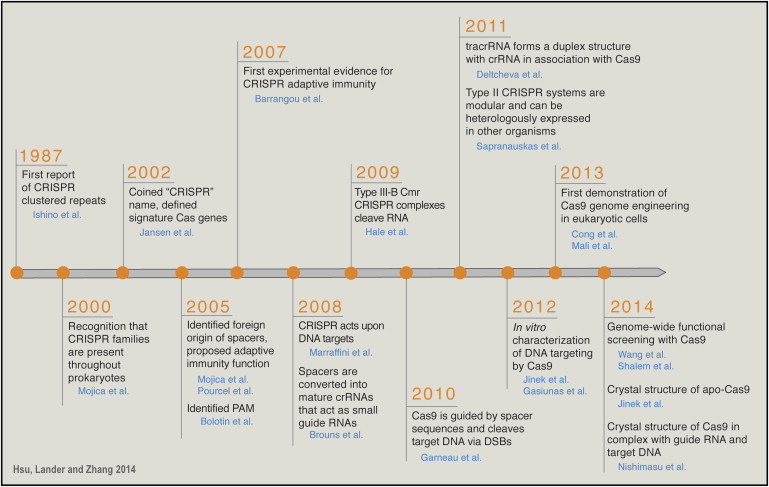
From adaptive immunity to genome editing
CRISPR/CAS genes were first found in bacteria [1] and archaea, enabling them to respond and eliminate invading genetic material (e.g. from virus or plasmids)[2].
As explained elegantly by Carl Zimmer:
As the CRISPR region fills with virus DNA, it becomes a molecular most-wanted gallery, representing the enemies the microbe has encountered. The microbe can then use this viral DNA to turn Cas enzymes into precision-guided weapons. The microbe copies the genetic material in each spacer into an RNA molecule. Cas enzymes then take up one of the RNA molecules and cradle it. Together, the viral RNA and the Cas enzymes drift through the cell. If they encounter genetic material from a virus that matches the CRISPR RNA, the RNA latches on tightly. The Cas enzymes then chop the DNA in two, preventing the virus from replicating.
Type II CRISPR system was most studied. Here foreign DNA is cut into small fragments and incorporated into a CRISPR locus amidst a series of short repeats. Transcripts of the CRISPR loci are then processed to generate small RNAs (crRNA), which are used to guide effector endonucleases targeting invading DNA based on sequence complementarity.
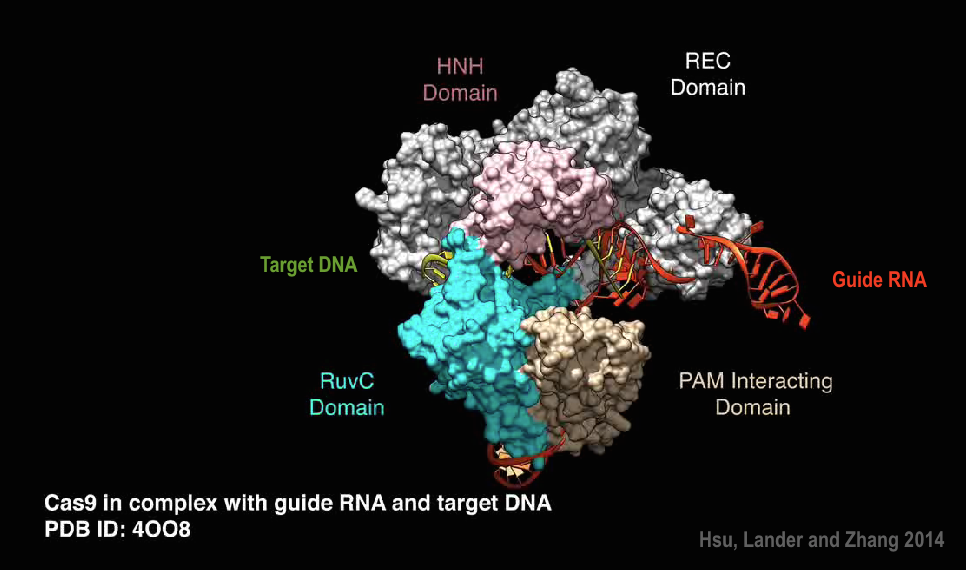
Type II CRISPR system has only three required components: Cas9 crRNA and trRNA, making it amenable to adaptation for genome editing. In 2012 a simplified two-component system was developed by combining trRNA and crRNA into a single synthetic single guide RNA (sgRNA), which can bind Cas9 and guide targeted gene alternation[3]. Scientists just need to modify the sequence of sgRNA to switch Cas9 to cut another DNA.
 {:hight=“325px” width=“550px”}
{:hight=“325px” width=“550px”}
In 2014 the CRISPR/Cas9 system was modified to a simplified genome-editing tool [4] also in eukaryotes, including yeast[5], C. elegans [6] and Drosophila [7], zebra fish, rats and even human cells[8].
More applications
Reversible knockdown
CRISPR interference (CRISPRi) using RNA-guided CRISPR associated nuclease Cas9 at the DNA level.
Gene activation
Using Cas9 to carry synthetic transcription factors to activate specific genes.
Imaging of specific genomic loci
Imaging location of genomic locus.[9]
Functional genomics
CRISPR/Cas9 enables rapid genome-wide interrogation of gene function by generating large gRNA libraries for genomic screening[10, 11].
FURTHER READING:
- CRISPR in the Lab: A Practical Guide
- How to Design Your gRNA for CRISPR Genome Editing
- Breakthrough DNA Editor Borne of Bacteria
- A CRISPR Fore-Cas-t
- Everything You Need to Know About CRISPR, the New Tool that Edits DNA
REFERENCES:
- Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product
- CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes
- A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity
- Development and Applications of CRISPR-Cas9 for Genome Engineering
- Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems
- Heritable genome editing in C. elegans via a CRISPR-Cas9 system
- Highly Improved Gene Targeting by Germline-Specific Cas9 Expression in Drosophila
- Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system
- CRISPR-Cas systems for editing, regulating and targeting genomes
- CRISPR/Cas9 and Targeted Genome Editing: A New Era in Molecular Biology
- High-throughput functional genomics using CRISPR–Cas9