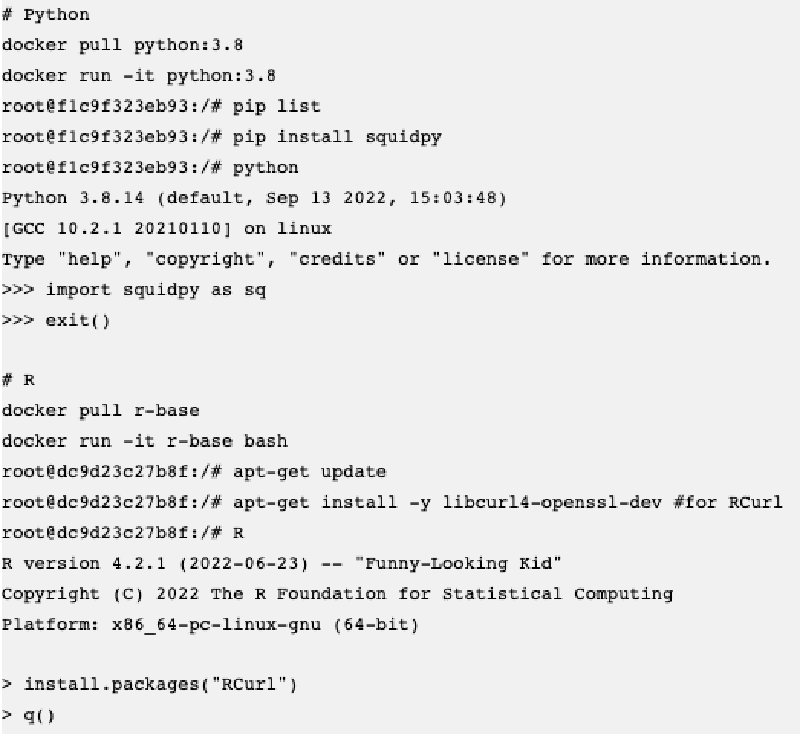
I am trying to build docker images with common tools for single-cell and spatial transcriptomics data analysis. The common way to build a new docker image is to use the Dockerfile, where we can define instructions and commands. For example here is one for R:
|
|
The problem is that many R packages have dependencies external to R, for example GSL, GDAL, JAGS and so on and we don’t know beforehand and difficult to define in the Dockerfile. To make a new image there is also a way to do it interactively using docker run -it [base image] bash which will bring us to the docker command line (like a system command line), and we can install system dependencies and further R/Python packages from >R or >python.
The good thing is that in R there will be indications what is needed:
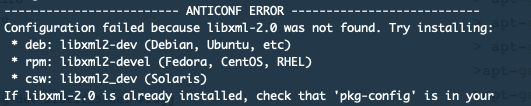
So just need to q() and go back to the command line and apt-get install
After we have installed all packages we can type docker ps in another terminal to see the running containers, and to make new image by typing docker commit [container id] image_name:tag
Then we can see the new image in docker images, which we can save and load in another system.
tested on my Mac
Other docker commands:
- docker push # to upload to dockerhub
- newgrp docker # after adding a user to the docker group