The information INPUT I have looks like this:
1
2
3
4
5
6
7
8
9
10
11
12
|
Smp_085010 SM_V7_1 3697949 PF00112,PF08127
Smp_168290 SM_V7_1 3714011
Smp_345790 SM_V7_1 3737539 PF00445
Smp_326260 SM_V7_1 3844720
Smp_103610 SM_V7_1 3867138 PF00112,PF08127
Smp_333870 SM_V7_1 3887599 PF00445
Smp_179960 SM_V7_1 3891316
Smp_333930 SM_V7_1 3908953 PF00445
Smp_333220 SM_V7_1 3925420 PF00445
Smp_334070 SM_V7_1 3968513 PF00445
Smp_334170 SM_V7_1 3980364 PF00445
Smp_334240 SM_V7_1 3992964 PF00445
|
I will number the genes according to their genomic positions (NrCoord, not considering of their strandness),
1
2
3
4
5
|
49 3697949 Smp_085010 PF00112,PF08127
50 3714011 Smp_168290
51 3737539 Smp_345790 PF00445
52 3844720 Smp_326260
53 3867138 Smp_103610 PF00112,PF08127
|
and split function terms for each gene, then to assign the gene orders to each function term (e.g. PF00445)
sort -k2,2 -k3,3n <INPUT> | awk '{print NR, $4}'| sed 's/,/ /g'| awk -v OFS='\t' '{for (i=2;i<=NF;i++) print $1,$i}'| awk '{print $2, $1}'| sort -k1,1 -k2,2n
Which looks like this:
1
2
3
4
5
6
7
8
|
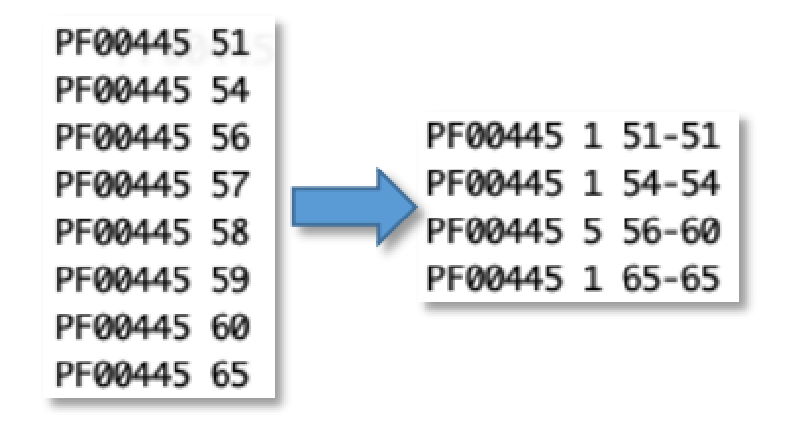
PF00445 51
PF00445 54
PF00445 56
PF00445 57
PF00445 58
PF00445 59
PF00445 60
PF00445 65
|
Then another awk to count consecutive numbers:
1
|
awk '$1>p || $2!=q+1{if(NR>1)print p,c,q-c+1,q; c=0} {p=$1; q=$2; c++} END{print p,c,q-c+1,q}'
|
to this: (the 56th to 60th genes all have PF00445)
1
2
3
4
|
PF00445 1 51 51
PF00445 1 54 54
PF00445 5 56 60
PF00445 1 65 65
|
Finally in R I can replace the gene orders ($3 and $4) with their coordinates from NrCoord, and get the list of genes in between those coordinates
1
2
3
4
5
6
7
8
9
10
11
|
#R example
for (i in 1:nrow(rawTable)) {
# replace gene order with gene coordinate
first<-rawTable[i, 4]
rawTable[i, 4]<-geneNrCoord[first, 2]
last<-rawTable[i, 5]
rawTable[i, 5]<-geneNrCoord[last, 2]
# get genes for the cluster
subgenes<-geneNrCoord[which(geneNrCoord$V2>=rawTable[i, 4] & geneNrCoord$V2<=rawTable[i, 5]),]
rawTable[i,6]<-paste(unlist(subgenes$V3), collapse=",")
}
|
to the table:
1
|
SM_V7_1 PF00445 Ribonuclease_T2 5 3908953 3992964 Smp_333930,Smp_333220,Smp_334070,Smp_334170,Smp_334240
|