This is a step-to-step guide. For more details please refer to the JBrowse documentation and the configuration guide.
To start with, Node.js is required to compile the necessary Perl scripts. Note that the installation of node.js requires admin.
Download and installation of JBrowse
I used the minified version, which doesn’t support plugins but is enough for essential functions.
|
|
Configure your JBrowse
Add your own genome sequence
|
|
When I used –indexed_fasta it didn’t work.
Add features / annotations and make them searchable
|
|
“GeneSet” is the appeared label for the annotation track. After generating names, we can search genes by their names.
Add other tracks from bed files
|
|
the bed file should contain the following columns: chr start end [name score strand] and score should be 0-1000
Note: the genome sequence, as well as the added tracks will go to the root folder “data/”, where:
|
|
Remove a flat file track
bin/remove-track.pl
But the script didn’t work in my hand. Need to remove the item from trackList.json and corresponding folder from tracks/.
Add a bam track
add the following to data/tracks.conf
|
|
Some tips:
To hide the strand arrow, add a line in trackList.json (don’t forget the “,” at the end of previous line)
|
|
To use the Faceted Track Selector, uncomment the following lines in “/jbrowse.conf”
|
|
Using the browser locally
After all the above configurations, we can simply open the index.html (recommend to use FireFox) and it will show your added genome and tracks. Bookmark that page for re-visting.
Hosting JBrowse on your own web server
We can build a JBrowse instance using free hosting services, e.g., Netlify. To do so, simply move the entire JBrowse-1.xxx folder into a new git repo on GitHub or Bitbucket, and deploy it from Netlify. Since there is an index.html in the JBrowser folder, the genome browser will be displayed on your website.
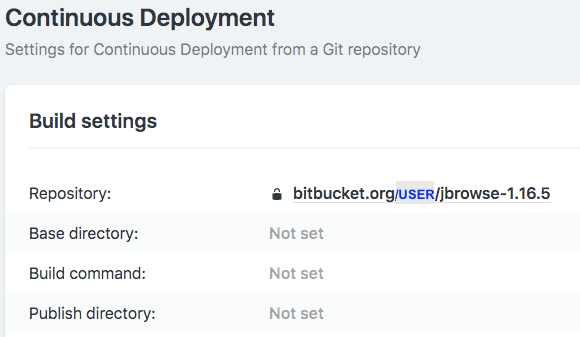
The advantage to use JBrowse on a web server is that it’s very easy to share the view of genomic locus by just sharing the url. And it’s easy to embed the view on any html page via ““iframe”. See below for an example