A few useful KEGG APIs
Get the reference names for K numbers
|
|
Get the names of reference pathways
|
|
- https://rest.kegg.jp/link/ko/map03013 (K orthologs in a map)
|
|
get K and ko links
|
|
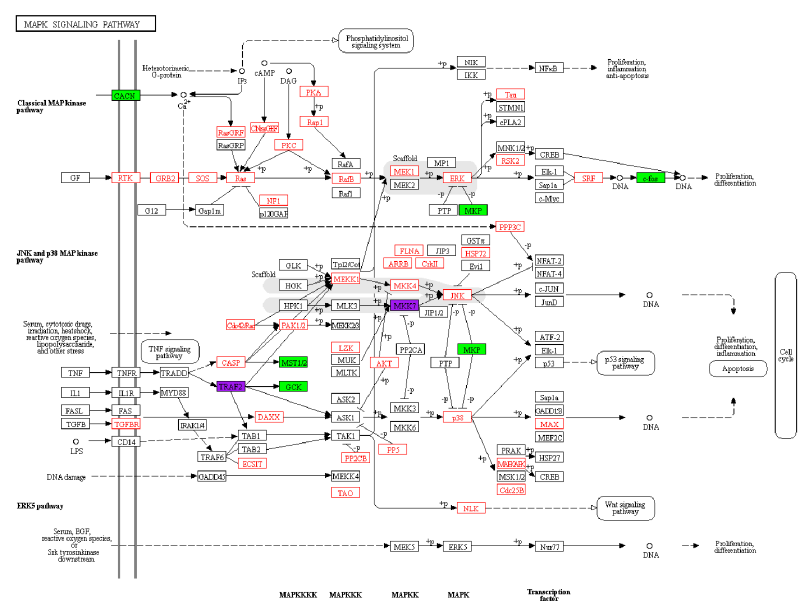
Pathway mapping and customisation
- https://www.genome.jp/kegg/mapper.html include various tools for mapping and coloring
or to open a specific pathway (with prefix “map” as initially no color shown, e.g. https://www.genome.jp/kegg-bin/show_pathway?map=map04010) and use “User data mapping”:
An input example
|
|
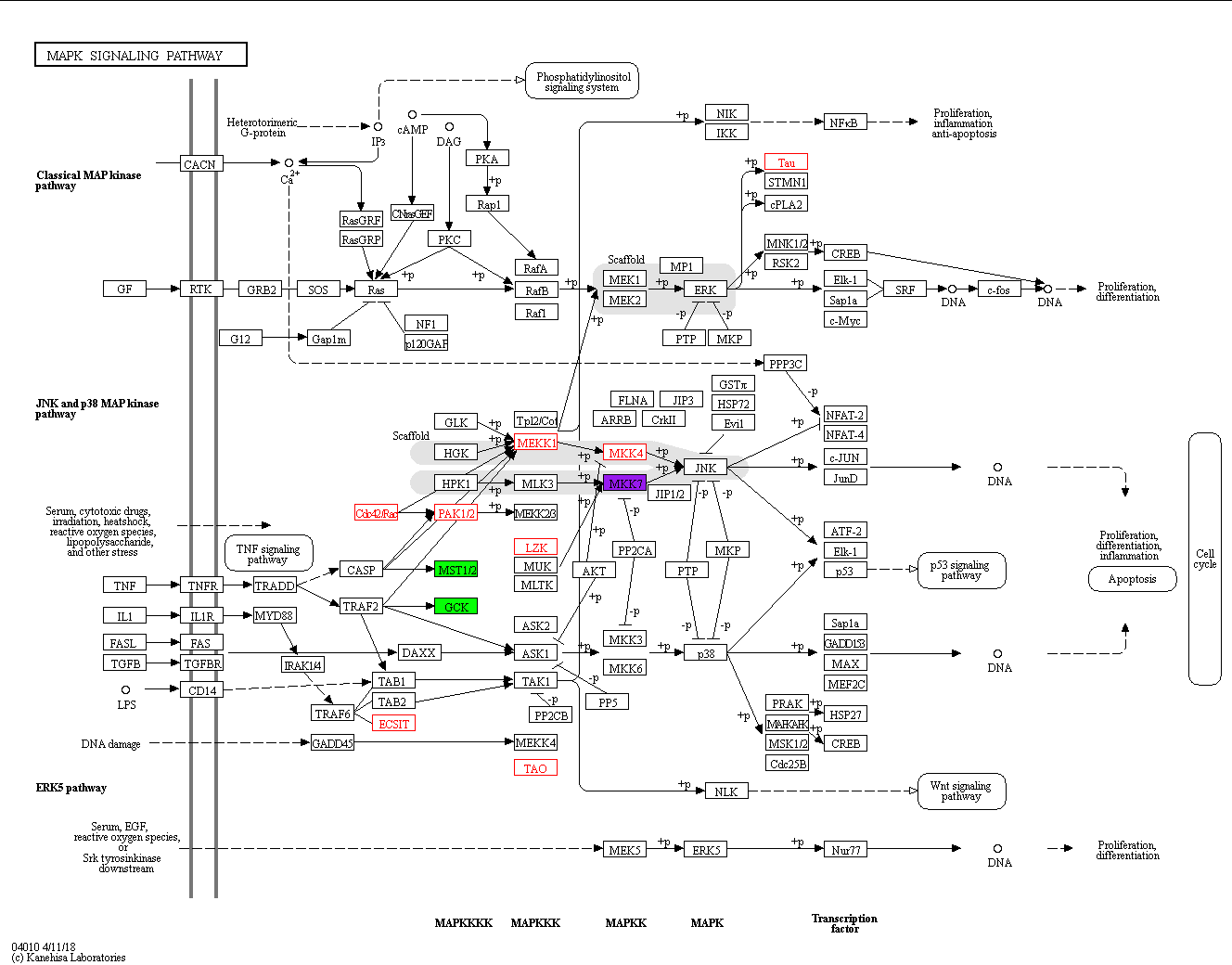
Or use the weblink api: https://www.kegg.jp/kegg/docs/weblink.html#map2
https://www.kegg.jp/kegg-bin/show_pathway?map=<mapid>&multi_query=<dataset>[&nocolor=1]